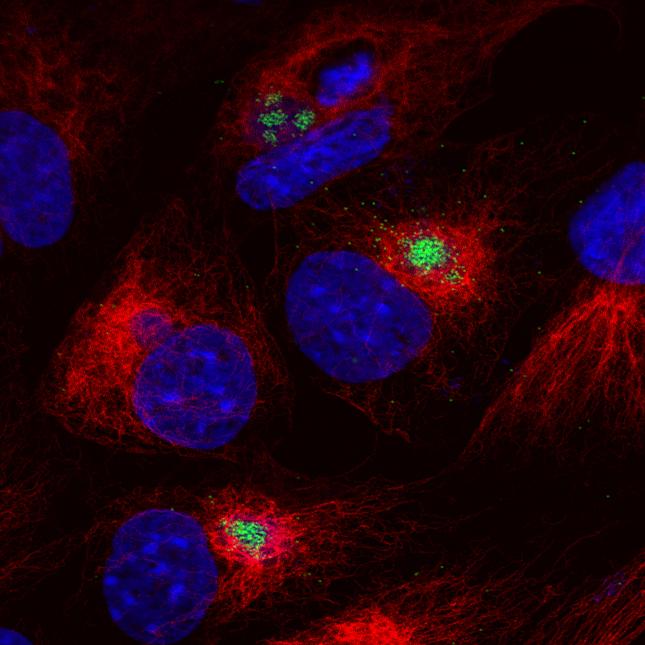
Transcription termination and readthrough in African swine fever virus
African swine fever virus (ASFV) is a nucleocytoplasmic large DNA virus (NCLDV) that encodes its own host-like RNA polymerase (RNAP) and factors required to produce mature mRNA. The formation of accurate mRNA 3′ ends by ASFV RNAP depends on transcription termination, likely enabled by a combination of sequence motifs and transcription factors, although these are poorly understood. The termination of any RNAP is rarely 100% efficient, and the transcriptional “readthrough” at terminators can generate long mRNAs which may interfere with the expression of downstream genes. ASFV transcriptome analyses reveal a landscape of heterogeneous mRNA 3′ termini, likely a combination of bona fide termination sites and the result of mRNA degradation and processing. While short-read sequencing (SRS) like 3′ RNA-seq indicates an accumulation of mRNA 3′ ends at specific sites, it cannot inform about which promoters and transcription start sites (TSSs) directed their synthesis, i.e., information about the complete and unprocessed mRNAs at nucleotide resolution.